
Epigenetic regulation by the Polycomb and Trithorax group proteins
Lab closed in 2014.
How do different cell types remember their identities over many cell generations? Part of the answer lies in the Polycomb and Trithorax groups of proteins. The Polycomb (PcG) and Trithorax (TrxG) groups of proteins work antagonistically on the same target genes, to maintain repressed (PcG) or active (TrxG) transcription states. We use a combination of quantitative live imaging, mathematical modelling, computational approaches and molecular and developmental biology to understand the interaction of the Polycomb and Trithorax proteins with their chromatin targets. We aim to unravel this fascinating epigenetic gene regulatory system in terms of the design, function and dynamic behaviour of its components. Our goal is to understand how a system whose components are in constant flux can ensure both stability and flexibility of gene expression states.
Leonie Ringrose has moved her lab to the IRI for Life Sciences at the Humboldt University, Berlin.
Research
Epigenetic regulation by the Polycomb and Trithorax group proteins
A single stem cell, with a single genomic DNA sequence, can give rise to an extraordinary diversity of cell identities and functions. The highly conserved Polycomb (PcG) and Trithorax (TrxG) group proteins constitute an epigenetic “cellular memory” system that is essential for maintaining the correct identity of both stem cells and differentiated cells. We aim to understand how this dynamic system can ensure both flexibility and stability of cell identities.

How does PRE/TRE regulation change during mitosis and differentiation?
The PcG and TrxG proteins act through Polycomb/Trithorax response elements (PRE/TREs). PRE/TREs are switchable bi-stable regulatory DNA elements that can preserve a memory of the activated or silenced state of their associated genes over several cell generations. We have previously developed an algorithm to predict fly PRE/TREs, and have examined the evolution of PRE/TRE elements across several Drosophila species, showing that PRE/TRE evolution is extraordinarily dynamic.
This year we have demonstrated an essential role in PRE/TRE switching for individual motifs that were predicted computationally, and have shown that different PRE/ TREs quantitatively modulate the output of their adjacent enhancer, showing for the first time that PRE/TREs from different genes and from different species have intrinsically different properties. Elucidating the molecular mechanisms by which PRE/TRE sequence contributes to gene activation, silencing and switching will be important tasks for the future.
In contrast to fly PRE/TREs, the corresponding mammalian elements have so far proved highly elusive. During the past year we have established reporter assays and training data sets for experimental and bioinformatic analysis of mammalian PRE/TREs. In future we hope to combine the expertise learned from the fly, with computational and experimental analysis in the mouse, to begin to tackle the question of what makes a mammalian PRE/TRE.
Noncoding RNAs in PRE/TRE regulation
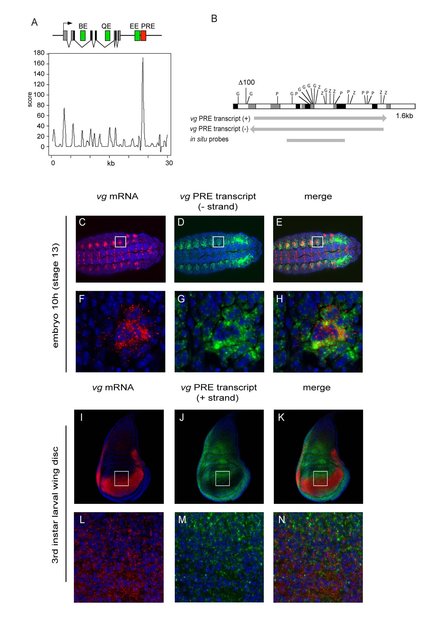
Our recent work in both fly and mouse has identified several novel long noncoding RNAs that are transcribed from polycomb regulatory sites, and suggests an essential role for these RNAs in PRE/TRE regulation during development and differentiation. In the fly, we have shown that the vestigial PRE/TRE undergoes a developmental switch, in which embryonic transcription of one PRE/TRE strand is associated with vestigial gene activation in specific cells, whereas larval transcription of the other strand is linked to gene repression (Figure 1).
In the mouse, analysis of purified cell populations from different stages of neural differentiation reveals sites of intergenic noncoding transcription in each cell type that precisely co-localize with PcG binding sites in the same or other cell types. Both of these studies have in common that they document a remarkable plasticity of transcription of these noncoding RNAs during development and differentiation. our future work will address the molecular mechanisms by which selected transcripts act on PcG/TrxG regulation in specific cell types and at specific developmental stages (Figure 2).
Quantitative live imaging and mathematical modeling
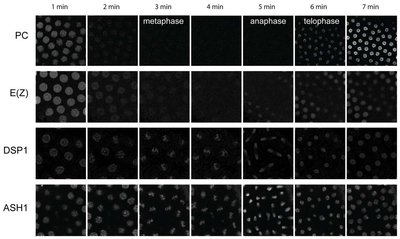
We have established an “in vivo biochemistry” approach to perform quantitative analysis of PcG and TrxG protein dynamics in living Drosophila in defined cell types that undergo mitosis and differentiation. This analysis shows fundamental differences in the chromatin binding properties of these proteins between stem cells and differentiated cells, and between different proteins (Figure 3).
This is the first study of an epigenetic system to quantify numbers of molecules and their kinetic constants during defined differentiation events in vivo. We have used the quantitative measurements to construct mathematical models that have identified parameters of the system that can explain the observed changes in chromatin binding plasticity upon mitosis and differentiation. In future we aim to elucidate the molecular mechanisms underlying these changes, and to determine their contribution to the establishment and maintenance of different cell identities.
Selected Publications
(2014)
Herzog, VA., Lempradl, A., Trupke, J., Okulski, H., Altmutter, C., Ruge, F., Boidol, B., Kubicek, S., Schmauss, G., Aumayr, K., Ruf, M., Pospisilik, A., Dimond, A., Senergin, HB., Vargas, ML., Simon, JA., Ringrose, L. (2014). A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet. 46(9):973-81
Steffen, PA., Ringrose, L. (2014). What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 15(5):340-56
(2013)
Steffen, PA., Fonseca, JP., Gänger, C., Dworschak, E., Kockmann, T., Beisel, C., Ringrose, L. (2013). Quantitative in vivo analysis of chromatin binding of Polycomb and Trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucleic Acids Res. 41(10):5235-50
(2012)
Fonseca, JP., Steffen, PA., Müller, S., Lu, J., Sawicka, A., Seiser, C., Ringrose, L. (2012). In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev. 26(8):857-71
Hekimoglu-Balkan, B., Aszodi, A., Heinen, R., Jaritz, M., Ringrose, L. (2012). Intergenic Polycomb target sites are dynamically marked by non-coding transcription during lineage commitment. RNA Biol. 9(3):314-25
Steffen, PA., Fonseca, JP., Ringrose, L. (2012). Epigenetics meets mathematics: towards a quantitative understanding of chromatin biology. Bioessays. 34(10):901-13
(2011)
Okulski, H., Druck, B., Bhalerao, S., Ringrose, L. (2011). Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics Chromatin. 4:4
(2009)
Hekimoglu, B., Ringrose, L. (2009). Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 6(2):129-37
(2008)
Hauenschild, A., Ringrose, L., Altmutter, C., Paro, R., Rehmsmeier, M. (2008). Evolutionary plasticity of polycomb/trithorax response elements in Drosophila species. PLoS Biol. 6(10):e261
Lempradl, A., Ringrose, L. (2008). How does noncoding transcription regulate Hox genes? Bioessays. 30(2):110-21