
As a lab, our goal aims to understand behaviors of adult stem cells in homeostasis and injury by focusing on three complementary aspects: maintenance, quiescence and developing genetic tools to uncover adult stem cells behavior in homeostasis. We utilize a variety of models for our studies as summarized in this graphical abstract illustrated by our current master’s student, Andreia Batista-Rocha.
Project I: Understanding the identity and characteristics of adult gastric stem cells
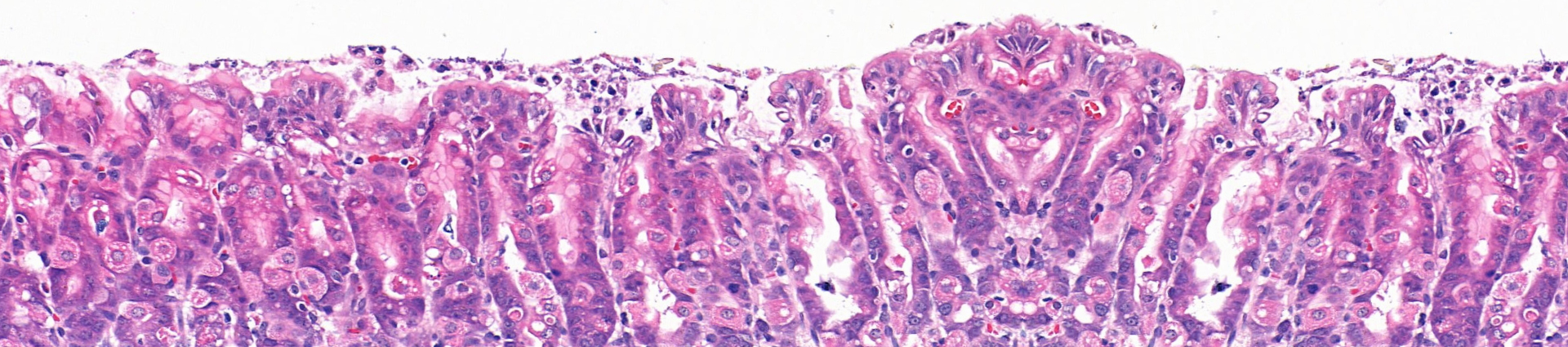
We previously identified Troy+ chief cells as a novel stem cell population in the corpus epithelium of the stomach (Stange and Koo et al, Cell, 2013). These cells proliferate slowly, indicating a rather quiescent nature compared to other known gastro-intestinal tract stem cells. The stomach corpus gland also contains a secondary stem cell population known as isthmus stem cells which actively cycles to maintain the pit-isthmus-neck region (Han et al, CSC, 2019). However, upon depletion of the isthmus stem cells, Troy+ cells are able to replenish the gland. This suggests distinct functions of these two stem cells under homeostasis and injury. As Troy+ stomach stem cells (StSCs) exhibit interchangeable characteristics, i.e. quiescent and proliferative, they represent a unique model of adult stem cells. This allows us to study:
- The dynamics of stem cell propagation in homeostasis and regeneration, as well as the underlying mechanism of this switch. Based on the changes in the mRNA expression and epigenetic profiles of Troy+ stem cells in homeostasis versus injury repair, we will generate a list of gene candidates that potentially play a role in cell fate decision. We will subsequently investigate the role of these candidates by using 3D organoid culture and mouse models.
- The StSC program in homeostasis and regeneration using in vitro and in vivo functional genetics. For this project, our team uses lineage tracing approaches with ubiquitous and cell type-specific CreERT2 transgenic mice, combined with diphtheria toxin receptor-mediated cell-type specific ablation models, tissue clarification, thick tissue imaging, and expression analysis at tissue, cell type-specific and single cell levels.
Project II: Analyzing the role of E3 Ubiquitin Ligases in the maintenance of adult stem cells
Homeostatic turnover in adult tissues is governed by the interplay of a multitude of signaling pathways. De-regulation of these pathways can result in pathogenic conditions, such as uncontrolled proliferation or stem cell depletion. Activation of these pathways is often triggered by niche cells, which provide diverse ligands to support the stem cells. Therefore, ligand-receptor interaction-initiated signaling cascades must be thoroughly and tightly controlled in order to sustain the functionality of the stem cells. An important class of such modulators is the E3 ubiquitin ligase family that specifically conjugate ubiquitin tags to their target proteins for ubiquitin-mediated degradation by proteasomes. Particularly, ubiquitination of membrane proteins directs trafficking decisions related to both biosynthetic delivery of proteins to the plasma membrane (PM) via the secretory pathway, and removal of proteins from the PM via the endocytic pathway.
Previously, Dr. Koo showed the crucial role of Mib1 (an E3 for Notch ligands) in Notch ligand activation in niche cells that, in turn, promoted Notch signaling in stem cells (Koo et al., Development, 2005; Koo et al., Gastroenterology, 2009). Dr. Koo also found that RNF43 and ZNRF3 attenuate Wnt activation in intestinal stem cells by functioning as E3s for Wnt receptors (Koo et al., Nature, 2012; Koo et al., PNAS, 2015). We aim at identifying other E3 ligases that play important roles in adult stem cell biology. For this project, three advanced technologies are combined to characterize the function of diverse E3 ligases playing an important role in tissue homeostasis.
- First, we utilize intestinal and gastric organoid cultures to test the effect of E3 ligase overexpression and knockout.
- Second, for selected E3 candidates, we perform extensive mass-spec analyses (e.g. surface proteome analysis, IP-Mass, and BIO-ID) to identify the substrate(s).
- Lastly, we have developed diverse genetic tools for organoid culture, enabling loss- and gain-of-function approaches in organoids.
Project III: Tracing the effect of oncogenic signals on adult stem cell behavior
Cancer develops through a multi-stage evolutionary process, proceeding from early meta-/neoplastic transformation to invasive carcinoma and metastasis. Current methods struggle to define mutation-containing early pre-tumorigenic lesions due to their phenotypic similarity with the surrounding normal tissue. We aim at defining the earliest steps of pre-neoplastic transformation, and identifying which tumorigenic mutations are the most threatening and require early medical intervention.
To advance this field of research, we developed novel variants of the Rosa-Confetti 4-colour reporter allele (Red2cDNA series). These variants harbor different oncogenes and site-specific recombinase (e.g. Flpe, for tumor suppressor knockout) coupled to a red fluorescent protein (RFP). Drawing upon the development of mathematical modelling methods, as well as advanced imaging and image-analysis, our team will define the clonal dynamics of tumorigenic mutation-containing clones. We will also evaluate the response of surrounding normal clones in homoeostasis and different tissue contexts, such as regeneration and inflammation. In this way, we can compare and contrast clonal dynamics in transformed mutant (in red) cells with neighboring normal clones (in green, yellow and cyan). This will allow us to:
- Investigate the clonal behavior of mutant clones in cell-to-cell competition in the tumor niche
- Probe the remodeling of the tumor niche and the involvement of non-mutated surrounding tissue in this process
- Analyze competition of large clones in the wide field of tissue.