HiCognition: A computational tool to navigate the genome in 3D
Complex functions such as gene expression, recombination, and chromosome segregation require highly regulated and coordinated folding of DNA inside cells. This organization depends on thousands of proteins and chemical modifications that determine chromatin structure. Yet, establishing a direct link between 3D genome organization and physiological functions is often complex and tedious, owing to the high variability between different genomic regions and the multitude of factors involved.
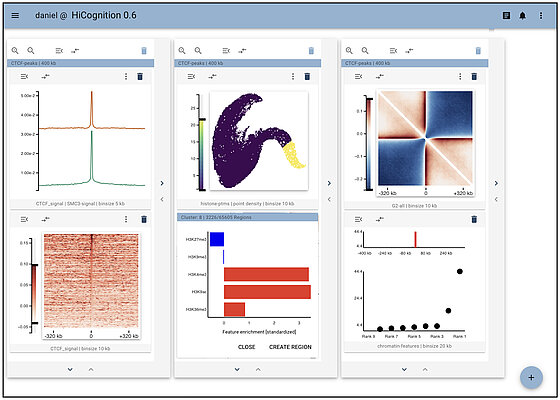
Researchers from IMBA Senior Scientist Daniel Gerlich’s lab developed HiCognition, a visualization and machine-learning tool that allows biologists to navigate large, multi-dimensional genomics data. According to the team, this tool will provide researchers with unprecedented opportunities to understand fundamental mechanisms that influence genome structure and function. The study, co-first authored by the bioinformaticians Christoph Langer and Michael Mitter, also provides an example of HiCognition’s applicability: the authors show how epigenetic marks as well as the cohesin complex organize the conformation of genes.
HiCognition combines an interactive visualization interface with high-performance data processing, statistical tools, and machine learning. HiCognition’s fast and computationally efficient implementation allows real-time browsing through thousands of genomic regions. These properties will substantially accelerate hypothesis testing by integrating data obtained with several experimental techniques, under various experimental conditions, and in different cell states.